About JAKSTAT TARGET
JAKSTAT-TARGET is an interdisciplinary consortium of immunologists, hematologists, structural chemists, systems biologists, and computational scientists. The consortium aims to investigate ways to improve individualized therapies for T-cell cancers by capitalizing on unique resources, such as clinical registry-linked sample repositories, new high-fidelity mouse models, pipelines of structure-based target design, and in-silico machine learning tools.
Background
Mature T-cell leukemias and lymphomas (MaTCL) are hematologic malignancies of mostly incurable prospects, as options for effective treatment are currently very limited. Furthermore, this heterogenous group of tumors poses challenges in large-volume biomaterial and data collections for clinical studies, since single MaTCL types are rare.
However, previous studies indicate that hyperactivating somatic mutations in JAK/STAT genes stand out as high-incidence aberrations across MaTCL entities. Particularly the model diseases investigated here, T-cell prolymphocytic leukemia (T-PLL) and T-cell large granular lymphocyte leukemia (T-LGLL), carry those somatic mutations in 50-70% of cases. This project proposes that targeting the JAK/STAT signalling network in synergistic combinations with drugs inhibiting other interconnected key driver pathways will improve individualized anti-leukemic efficacy.
Timeline
During this 36-month project, the consortium aims to complete 3 objectives as illustrated in the figure below:
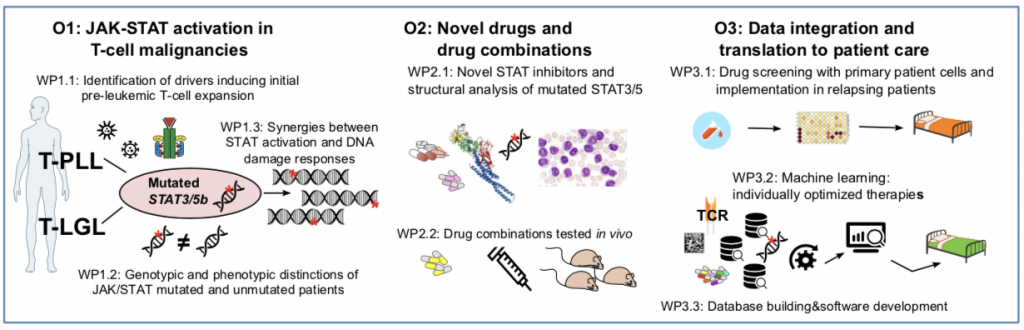
- O1 will address the causal T-cell-receptor and cytokine mediated impact on genome integrity and on the occurrence of JAK/STAT mutations. It interrogates the biochemical and functional consequences in the mutated and unmutated clones, and derives differential actionable vulnerabilities.
- O2 will study the performance of identified synergistic drug combinations using in-house developed optimized STAT-inhibitors in primary patient samples and novel animal models
- O3 will implement drug-screen data from patient material into an ongoing clinical trial.
Ultimately, with machine-learning algorithms we will integrate the harmonized data of genomic profiles, drug-sensitivity patterns, and clinical outcomes from all objectives toward multi-omics guided predictions of optimal choices for trial designs and individual-patient based therapy selection.



